Conversion from CellRank pipeline¶
The aim of this notebook is to convert resulting analysis from CellRank into a principal tree that can be used by scFates
CellRank aims at identifying fate potentials by considering single cell dynamics as a Markov process (see Lange et al., biorxiv, 2021). This is a great tool for finding the macrostates such as the “tips” of our trajectories, thanks to its powerful probabilistic approach. Here we propose to extend it by converting the fate probabilities into a principal tree, allowing easier interpretation of what is happening “in between” (early biases, bifurcations).
Setting up environment modules and basic settings¶
Generating the environment¶
The following needs to be run in the command-line
conda create -n scFates -c conda-forge -c r python=3.8 r-mgcv rpy2=3.4.2 -y
conda activate scFates
# Install new jupyter server
conda install -c conda-forge jupyter
# Or add to an existing jupyter server
conda install -c conda-forge ipykernel
python -m ipykernel install --user --name scFates --display-name "scFates"
# Install scFates
pip install scFates
Required additional packages¶
[1]:
import sys
!{sys.executable} -m pip -q install scvelo cellrank
Loading modules and settings¶
[2]:
import scvelo as scv
import scanpy as sc
import cellrank as cr
import numpy as np
scv.settings.verbosity = 3
scv.settings.set_figure_params('scvelo')
cr.settings.verbosity = 2
[3]:
import warnings
warnings.simplefilter("ignore", category = UserWarning)
warnings.simplefilter("ignore", category = FutureWarning)
Reproduction of CellRank notebook¶
Here we run a compressed version of the CellRank notebook which reproduces figure 2 of their paper.
[4]:
adata = cr.datasets.pancreas()
adata.var_names.name=None
adata.raw = adata # We want to keep all the genes for testing on the resulting tree
scv.pp.filter_genes(adata,min_shared_counts=20)
scv.pp.filter_and_normalize(adata, min_shared_counts=20, n_top_genes=2000)
sc.tl.pca(adata)
sc.pp.neighbors(adata, n_pcs=30, n_neighbors=30)
scv.pp.moments(adata, n_pcs=None, n_neighbors=None)
scv.tl.recover_dynamics(adata, n_jobs=20)
scv.tl.velocity(adata, mode='dynamical')
scv.tl.velocity_graph(adata)
scv.tl.latent_time(adata)
Filtered out 22024 genes that are detected 20 counts (shared).
Normalized count data: X, spliced, unspliced.
Extracted 2000 highly variable genes.
Logarithmized X.
computing moments based on connectivities
finished (0:00:00) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
recovering dynamics (using 20/88 cores)
WARNING: Unable to create progress bar. Consider installing `tqdm` as `pip install tqdm` and `ipywidgets` as `pip install ipywidgets`,
or disable the progress bar using `show_progress_bar=False`.
finished (0:00:38) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
computing velocities
finished (0:00:02) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
computing velocity graph (using 1/88 cores)
finished (0:00:04) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
computing terminal states
WARNING: Uncertain or fuzzy root cell identification. Please verify.
identified 1 region of root cells and 1 region of end points .
finished (0:00:00) --> added
'root_cells', root cells of Markov diffusion process (adata.obs)
'end_points', end points of Markov diffusion process (adata.obs)
computing latent time using root_cells as prior
finished (0:00:00) --> added
'latent_time', shared time (adata.obs)
[5]:
from cellrank.tl.estimators import GPCCA
weight_connectivities=0.2
mode="stochastic"
n_jobs=40
softmax_scale=None
kernel = cr.tl.transition_matrix(adata,
weight_connectivities=weight_connectivities,
mode=mode,
n_jobs=n_jobs,
softmax_scale=softmax_scale)
g_fwd = GPCCA(kernel)
g_fwd.compute_schur(n_components=20)
n_states = 12
g_fwd.compute_macrostates(cluster_key='clusters', n_states=n_states)
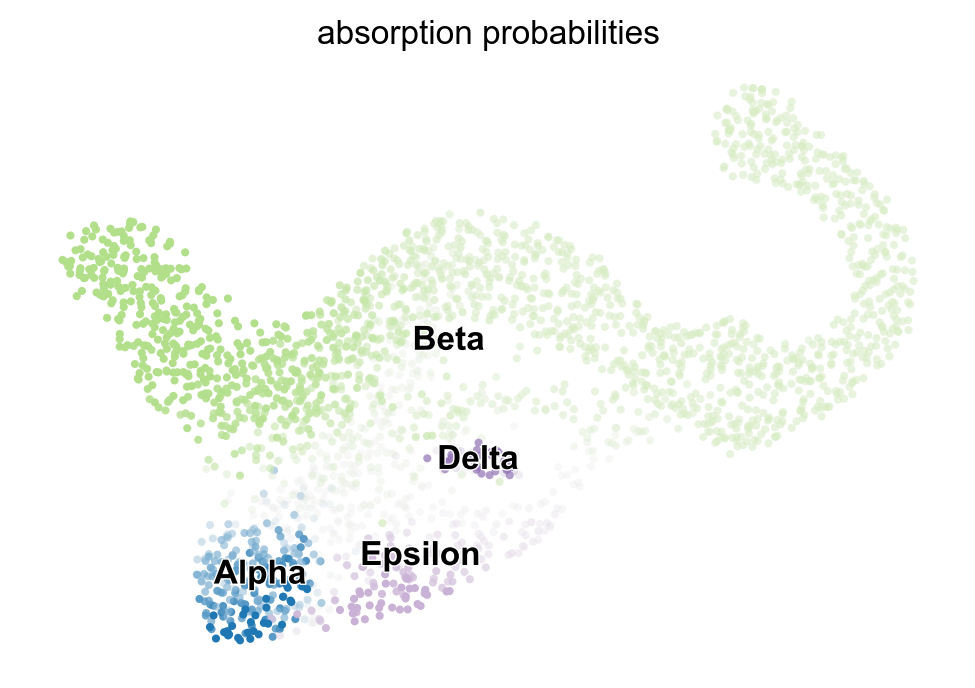
g_fwd.set_terminal_states_from_macrostates(names=['Alpha', 'Beta', 'Epsilon', 'Delta'])
g_fwd.compute_absorption_probabilities()
cr.pl.lineages(adata)
WARNING: Unable to detect a method for Hessian computation. If using predefined functions, consider installing `jax` as `pip install jax jaxlib`
Defaulting to `mode='monte_carlo'` and `n_samples=1000`
Computing transition matrix based on logits using `'monte_carlo'` mode
/tmp/ipykernel_1233105/1773602998.py:7: DeprecationWarning: `cellrank.tl.transition_matrix` will be removed in version `2.0`. Please use the `cellrank.kernels` or `cellrank.estimators` interface instead.
kernel = cr.tl.transition_matrix(adata,
Estimating `softmax_scale` using `'deterministic'` mode
100%|██████████| 2531/2531 [00:05<00:00, 429.55cell/s]
Setting `softmax_scale=3.7951`
100%|██████████| 2531/2531 [00:20<00:00, 122.11sample/s]
Finish (0:00:27)
Using a connectivity kernel with weight `0.2`
Computing transition matrix based on `adata.obsp['connectivities']`
Finish (0:00:00)
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
Computing Schur decomposition
When computing macrostates, choose a number of states NOT in `[6, 9, 15, 17]`
Adding `adata.uns['eigendecomposition_fwd']`
`.schur_vectors`
`.schur_matrix`
`.eigendecomposition`
Finish (0:00:07)
Computing `12` macrostates
Adding `.macrostates`
`.macrostates_memberships`
`.coarse_T`
`.coarse_initial_distribution
`.coarse_stationary_distribution`
`.schur_vectors`
`.schur_matrix`
`.eigendecomposition`
Finish (0:00:18)
Adding `adata.obs['terminal_states']`
`adata.obs['terminal_states_probs']`
`.terminal_states`
`.terminal_states_probabilities`
`.terminal_states_memberships
Finish`
Computing absorption probabilities
WARNING: Unable to import petsc4py. For installation, please refer to: https://petsc4py.readthedocs.io/en/stable/install.html.
Defaulting to `'gmres'` solver.
100%|██████████| 4/4 [00:00<00:00, 38.09/s]
Adding `adata.obsm['to_terminal_states']`
`.absorption_probabilities`
Finish (0:00:00)
Converting Cellrank output into a principal tree¶
[6]:
warnings.simplefilter("ignore", category = DeprecationWarning)
import scFates as scf
[7]:
scf.tl.cellrank_to_tree(adata,time="latent_time",Nodes=300,seed=1)
Converting CellRank results to a principal tree --> with .obsm['X_fates'], created by combining:
.obsm['X_fate_simplex_fwd'] (from cr.pl.circular_projection) and adata.obs['latent_time']
Solving TSP for `4` states
inferring a principal tree inferring a principal tree --> parameters used
300 principal points, sigma = 0.1, lambda = 100, metric = euclidean
fitting: 54%|█████▍ | 27/50 [00:02<00:02, 9.58it/s]
converged
finished (0:00:02) --> added
.uns['ppt'], dictionnary containing inferred tree.
.obsm['X_R'] soft assignment of cells to principal points.
.uns['graph']['B'] adjacency matrix of the principal points.
.uns['graph']['F'] coordinates of principal points in representation space.
finished (0:00:02) .obsm['X_fates'] representation used for fitting the tree.
.uns['graph']['pp_info'].time has been updated with latent_time
.uns['graph']['pp_seg'].d has been updated with latent_time
[8]:
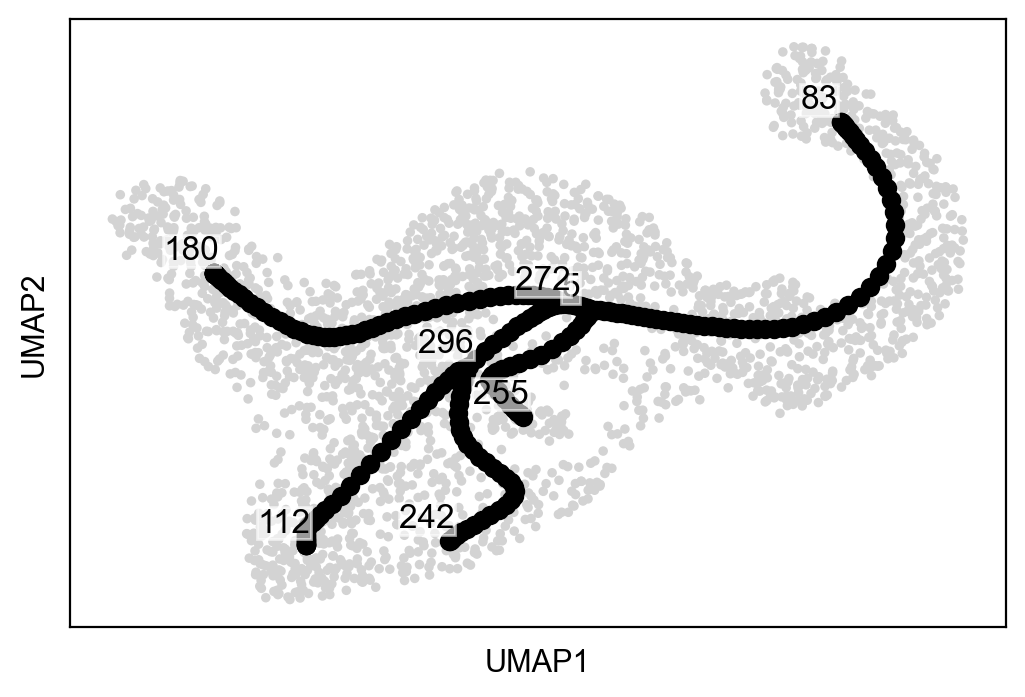
scf.pl.graph(adata)
[9]:
scf.tl.root(adata,83)
scf.tl.pseudotime(adata,n_jobs=20,n_map=100,seed=42)
node 83 selected as a root --> added
.uns['graph']['root'] selected root.
.uns['graph']['pp_info'] for each PP, its distance vs root and segment assignment.
.uns['graph']['pp_seg'] segments network information.
projecting cells onto the principal graph
mappings: 100%|██████████| 100/100 [00:36<00:00, 2.75it/s]
finished (0:00:38) --> added
.obs['edge'] assigned edge.
.obs['t'] pseudotime value.
.obs['seg'] segment of the tree assigned.
.obs['milestones'] milestone assigned.
.uns['pseudotime_list'] list of cell projection from all mappings.
[10]:
sc.pl.umap(adata,color=["seg","milestones"])
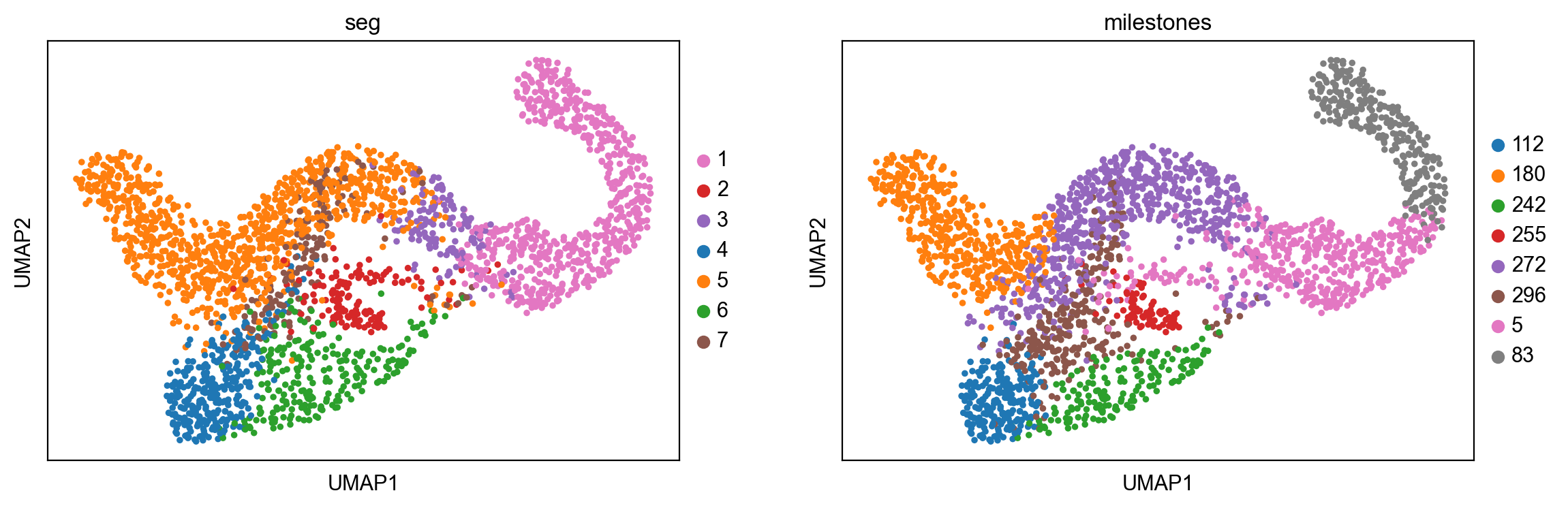
[11]:
# to avoid overlapping of labels, we set some intermediate states to empty strings of varying length
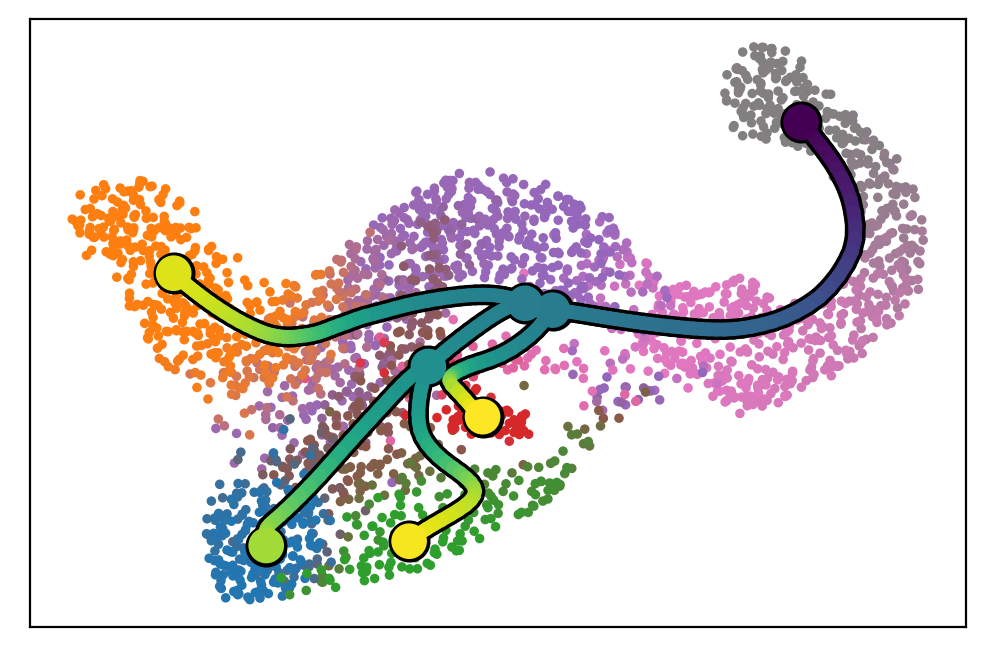
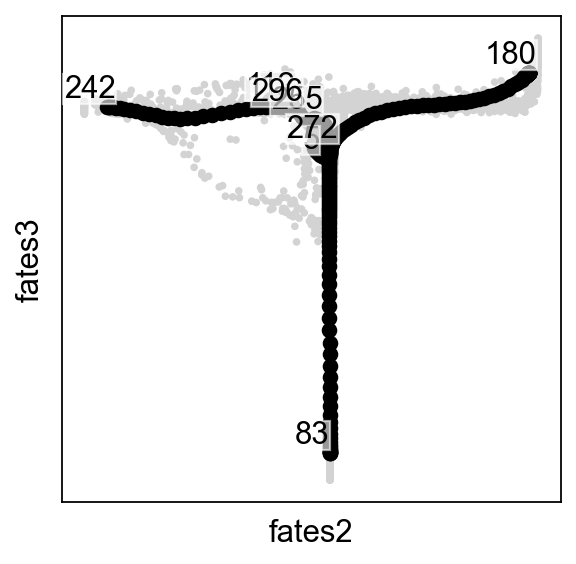
dct={"112":"Alpha","180":"Beta","5":"Ngn3 high EP","242":"Epsilon","255":"Delta","272":" ","296":" ","83":"Ngn3 low EP"}
[12]:
scf.tl.rename_milestones(adata,dct)
[13]:
scf.pl.trajectory(adata,color_cells="milestones")
[14]:
scf.tl.dendrogram(adata)
Generating dendrogram of tree
segment : 100%|██████████| 7/7 [00:01<00:00, 4.07it/s]
finished (0:00:01) --> added
.obsm['X_dendro'], new embedding generated.
.uns['dendro_segments'] tree segments used for plotting.
[15]:
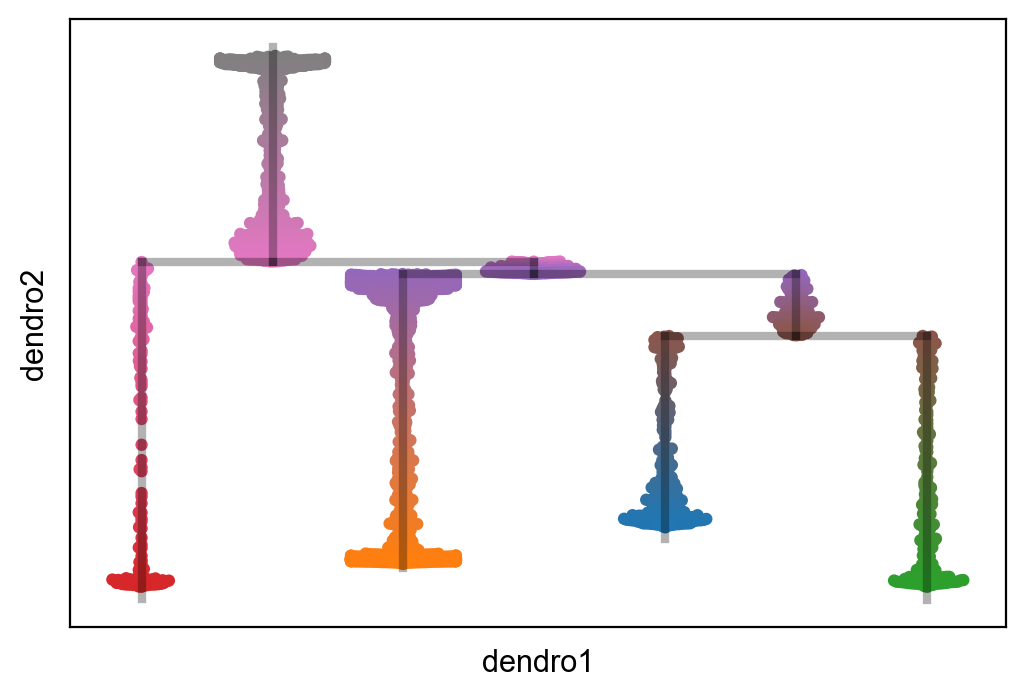
scf.pl.dendrogram(adata,color_milestones=True)
[16]:
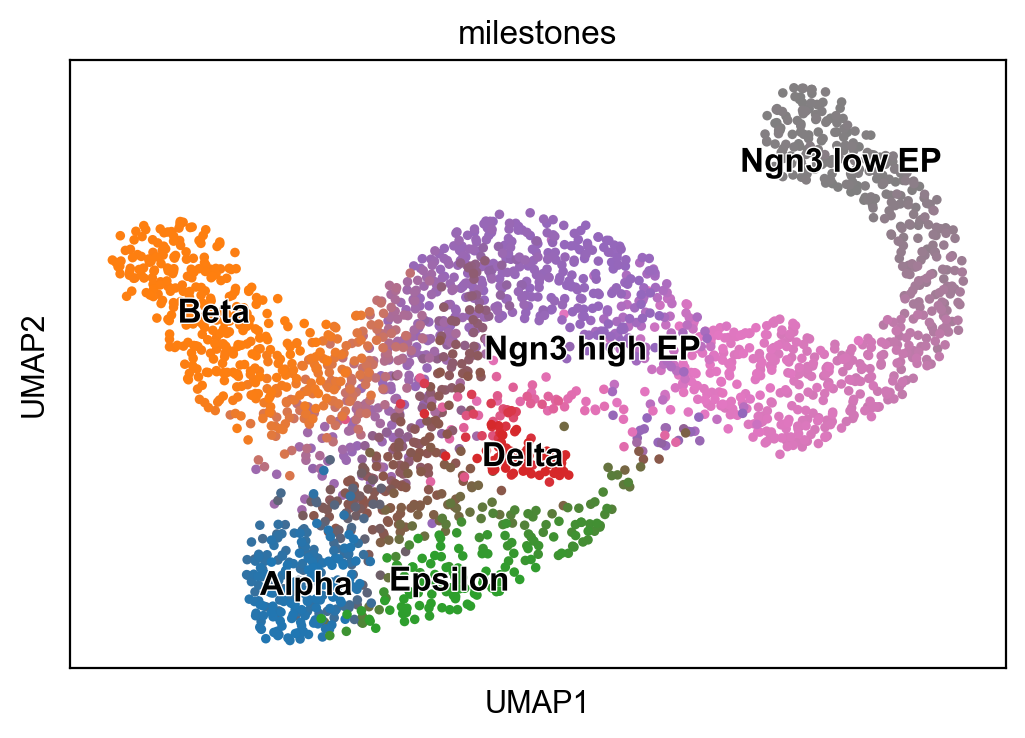
scf.pl.milestones(adata,annotate=True)
How the conversion is performed¶
Note
The following section is not needed for your own analysis pipeline, it is rather an explanation of what is going on when running the function scf.tl.cellrank_to_tree
From the scvelo/CellRank pipeline, we used the latent time estimate:
[17]:
scv.tl.latent_time(adata)
computing latent time using root_cells as prior
finished (0:00:00) --> added
'latent_time', shared time (adata.obs)
And a projection of the fate probabilities:
[18]:
cr.pl.circular_projection(adata,"clusters",legend_loc="right")
Solving TSP for `4` states
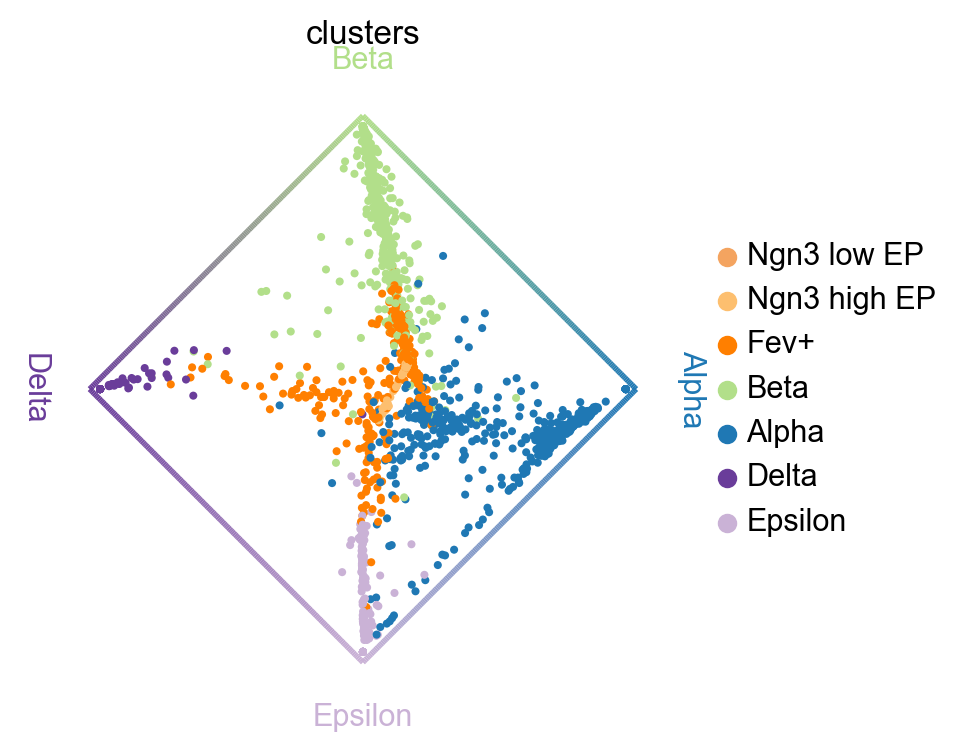
This projection generated bycr.pl.circular_projection can be found under:
[19]:
adata.obsm["X_fate_simplex_fwd"]
[19]:
array([[ 0.17996252, 0.13099097],
[ 0.27331319, -0.11478642],
[ 0.17542952, 0.09987978],
...,
[ 0.15714817, 0.19592445],
[ 0.83724649, -0.12325167],
[ 0.00368501, -0.80083918]])
if we add a third dimension using our latent time or any pseudotime measurement, we obtain the following
[20]:
adata.obsm["X_fates"]=np.concatenate([adata.obsm["X_fate_simplex_fwd"],
adata.obs["latent_time"].values.reshape(-1,1)],axis=1)
[21]:
adata.obsm["X_fates"]
[21]:
array([[ 0.17996252, 0.13099097, 0.8600946 ],
[ 0.27331319, -0.11478642, 0.88931465],
[ 0.17542952, 0.09987978, 0.65183399],
...,
[ 0.15714817, 0.19592445, 0.82558667],
[ 0.83724649, -0.12325167, 0.85034744],
[ 0.00368501, -0.80083918, 0.83946749]])
[22]:
sc.pl.embedding(adata,basis="fates",projection="3d",color=["clusters","latent_time"],cmap="gnuplot")

We then fitted a principal tree in this 3d space! Given that each dimension is between 0 and 1, scFates should be able to easily capture all the tips without having to tweak the parameters.
[23]:
sc.set_figure_params()
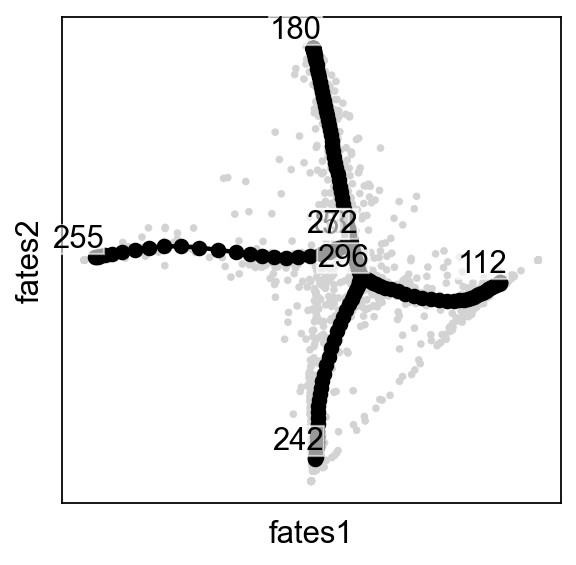
scf.pl.graph(adata,basis="fates")
scf.pl.graph(adata,basis="fates",dimensions=[1,2])
/home/lfaure/miniconda3/envs/scFates/lib/python3.8/site-packages/scanpy/_settings.py:447: DeprecationWarning: `set_matplotlib_formats` is deprecated since IPython 7.23, directly use `matplotlib_inline.backend_inline.set_matplotlib_formats()`
IPython.display.set_matplotlib_formats(*ipython_format)
What about the case of two terminal states only?
No simplex representation can be generated, in such case, one column from adata.obsm['to_terminal_states'] is used as 2d representation to be combined with the time estimate.
What about multiple initial states?
We could combine adata.obsm['X_fate_simplex_fwd'] or adata.obsm['to_terminal_states'] with adata.obsm['X_fate_simplex_bwd'] or adata.obsm['from_intial_states'] into a 4d representation, this will be implemented and tested in a future version.
Downstream analysis¶
Test and fitting associated genes¶
[24]:
adata=adata.raw.to_adata()
[25]:
sc.pp.filter_genes(adata,min_cells=3)
sc.pp.normalize_total(adata,target_sum=1e6)
sc.pp.log1p(adata,base=10)
[26]:
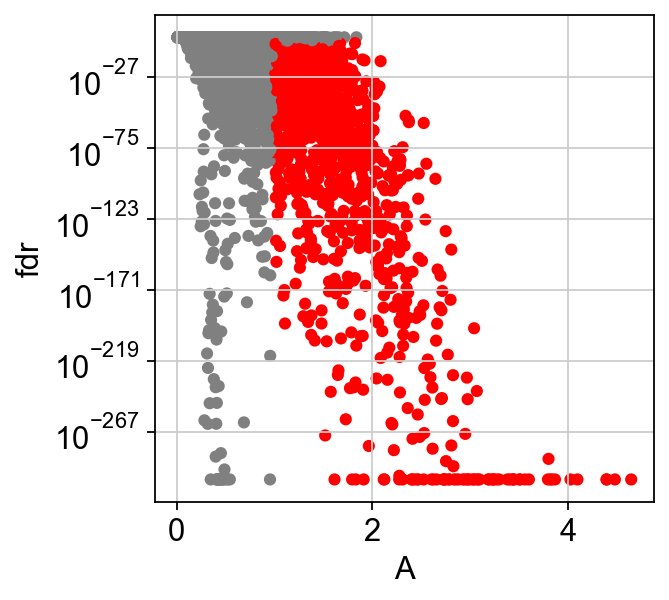
scf.tl.test_association(adata,n_jobs=20)
test features for association with the trajectory
single mapping : 100%|██████████| 14939/14939 [02:34<00:00, 96.67it/s]
found 1545 significant features (0:02:34) --> added
.var['p_val'] values from statistical test.
.var['fdr'] corrected values from multiple testing.
.var['st'] proportion of mapping in which feature is significant.
.var['A'] amplitue of change of tested feature.
.var['signi'] feature is significantly changing along pseudotime.
.uns['stat_assoc_list'] list of fitted features on the graph for all mappings.
[27]:
scf.pl.test_association(adata)
[28]:
scf.tl.fit(adata,n_jobs=20)
fit features associated with the trajectory
single mapping : 100%|██████████| 1545/1545 [00:45<00:00, 34.11it/s]
finished (adata subsetted to keep only fitted features!) (0:00:46) --> added
.layers['fitted'], fitted features on the trajectory for all mappings.
.raw, unfiltered data.
Identification of branch specific genes¶
[29]:
root='Ngn3 low EP'
miles=['Alpha','Epsilon','Beta','Delta']
[30]:
scf.tl.test_fork(adata,root_milestone=root,milestones=miles,n_jobs=20,rescale=True)
testing fork
single mapping
Differential expression: 100%|██████████| 1545/1545 [00:25<00:00, 61.35it/s]
test for upregulation for each leave vs root
upreg Alpha: 100%|██████████| 397/397 [00:00<00:00, 657.44it/s]
upreg Epsilon: 100%|██████████| 480/480 [00:00<00:00, 496.55it/s]
upreg Beta: 100%|██████████| 383/383 [00:00<00:00, 591.96it/s]
upreg Delta: 100%|██████████| 285/285 [00:00<00:00, 882.93it/s]
finished (0:00:28) --> added
.uns['Ngn3 low EP->Alpha<>Epsilon<>Beta<>Delta']['fork'], DataFrame with fork test results.
[31]:
scf.tl.branch_specific(adata,root_milestone=root,milestones=miles,effect=.3)
branch specific features: Alpha: 53, Beta: 44, Delta: 8, Epsilon: 7
finished --> updated
.uns['Ngn3 low EP->Alpha<>Epsilon<>Beta<>Delta']['fork'], DataFrame updated with additionnal 'branch' column.
[32]:
adata.obs_names.name=None
[33]:
from matplotlib import MatplotlibDeprecationWarning
warnings.simplefilter("ignore", category = MatplotlibDeprecationWarning)
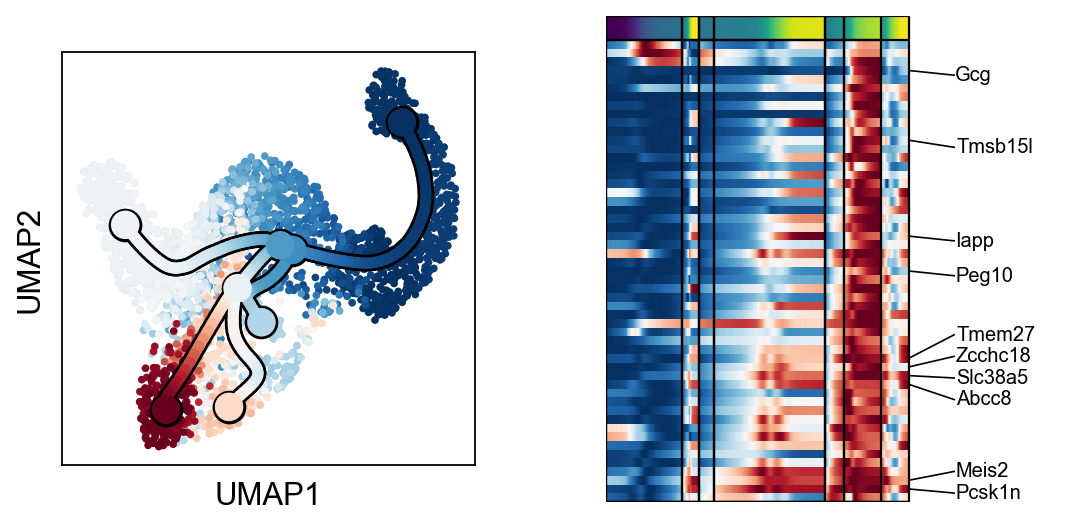
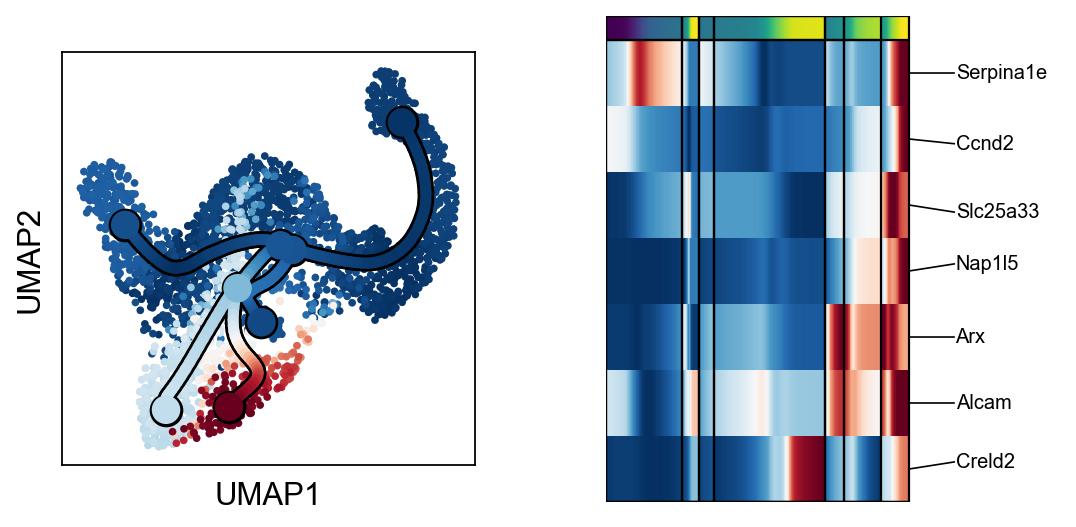
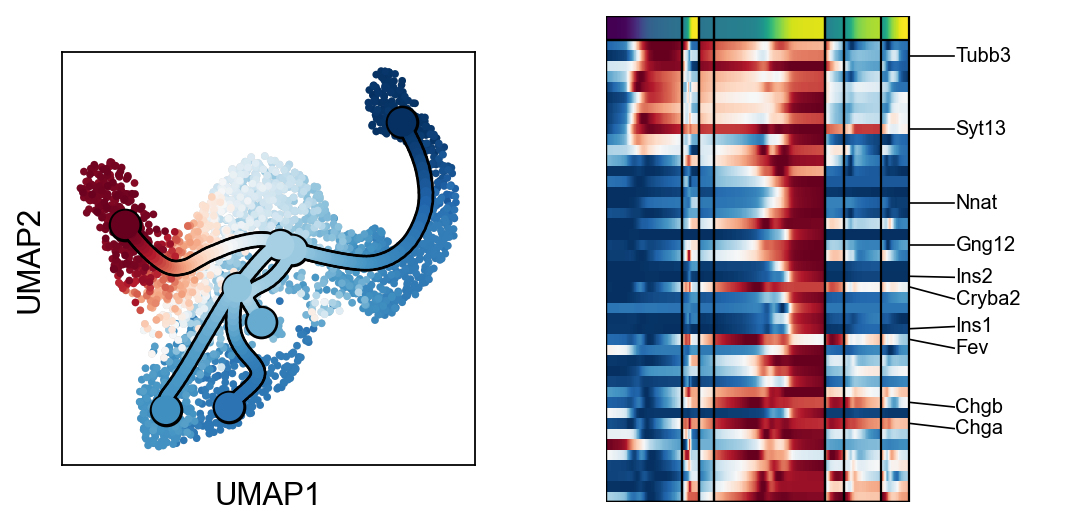
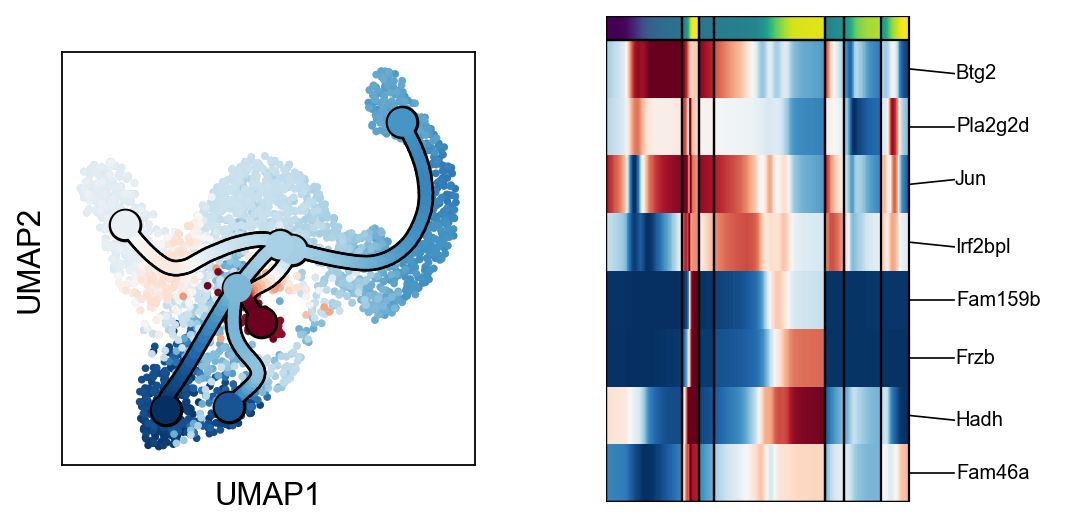
scf.pl.trends(adata,root_milestone=root,milestones=miles,branch=miles[0])
scf.pl.trends(adata,root_milestone=root,milestones=miles,branch=miles[1])
scf.pl.trends(adata,root_milestone=root,milestones=miles,branch=miles[2])
scf.pl.trends(adata,root_milestone=root,milestones=miles,branch=miles[3])